MLlib1.6指南笔记
http://spark.apache.org/docs/latest/mllib-guide.html
- spark.mllib RDD之上的原始API
- spark.ml ML管道结构 DataFrames之上的高级API
1. spark.mllib:数据类型、算法及工具
cd /Users/erichan/garden/spark-1.6.0-bin-hadoop2.6/bin./spark-shell --master local --driver-memory 6g
1.1 数据类型
1 局部向量(Local vector)
- 密集向量(dense)double数组
- 稀疏向量(sparse)两个平行数组:索引、值
Vector dv = Vectors.dense(1.0, 0.0, 3.0); Vector sv = Vectors.sparse(3, new int[] {0, 2}, new double[] {1.0, 3.0}); 2 标记点(Labeled point)
用于有监督学习算法(回归、分类)的局部向量。
LabeledPoint pos = new LabeledPoint(1.0, Vectors.dense(1.0, 0.0, 3.0)); LabeledPoint neg = new LabeledPoint(0.0, Vectors.sparse(3, new int[] {0, 2}, new double[] {1.0, 3.0})); LIBSVM格式
label index1:value1 index2:value2 ... JavaRDD<LabeledPoint> examples = MLUtils.loadLibSVMFile(jsc.sc(), "data/mllib/sample_libsvm_data.txt").toJavaRDD(); 3 局部矩阵(Local matrix)
- 密集矩阵(DenseMatrix)一维数组 列优先
- 稀疏矩阵(SparseMatrix)
Matrix dm = Matrices.dense(3, 2, new double[] {1.0, 3.0, 5.0, 2.0, 4.0, 6.0}); Matrix sm = Matrices.sparse(3, 2, new int[] {0, 1, 3}, new int[] {0, 2, 1}, new double[] {9, 6, 8}); 4 分布式矩阵(Distributed matrix)
行矩阵(RowMatrix) 每行是一个局部向量
JavaRDD<Vector> rows = ... //局部向量 JavaRDD RowMatrix mat = new RowMatrix(rows.rdd()); long m = mat.numRows(); long n = mat.numCols(); // QR分解 QRDecomposition<RowMatrix, Matrix> result = mat.tallSkinnyQR(true); 索引行矩阵(IndexedRowMatrix)每行是一个长整型和一个局部向量
JavaRDD<IndexedRow> rows = ... IndexedRowMatrix mat = new IndexedRowMatrix(rows.rdd()); long m = mat.numRows(); long n = mat.numCols(); // 去掉行索引 成为行矩阵 RowMatrix rowMat = mat.toRowMatrix(); 坐标矩阵(CoordinateMatrix) 行 列 值
JavaRDD<MatrixEntry> entries = ... CoordinateMatrix mat = new CoordinateMatrix(entries.rdd()); long m = mat.numRows(); long n = mat.numCols(); // Convert it to an IndexRowMatrix whose rows are sparse vectors. IndexedRowMatrix indexedRowMatrix = mat.toIndexedRowMatrix(); 分块矩阵(BlockMatrix) 索引元组 子矩阵
JavaRDD<MatrixEntry> entries = ... // a JavaRDD of (i, j, v) Matrix Entries // Create a CoordinateMatrix from a JavaRDD<MatrixEntry>. CoordinateMatrix coordMat = new CoordinateMatrix(entries.rdd()); // Transform the CoordinateMatrix to a BlockMatrix BlockMatrix matA = coordMat.toBlockMatrix().cache(); // Validate whether the BlockMatrix is set up properly. Throws an Exception when it is not valid. // Nothing happens if it is valid. matA.validate(); // Calculate A^T A. BlockMatrix ata = matA.transpose().multiply(matA); 1.2 统计
1 摘要统计
JavaRDD<Vector> mat = ... MultivariateStatisticalSummary summary = Statistics.colStats(mat.rdd()); System.out.println(summary.mean()); System.out.println(summary.variance()); System.out.println(summary.numNonzeros()); 2 相关统计
JavaSparkContext jsc = ... JavaDoubleRDD seriesX = ... // a series JavaDoubleRDD seriesY = ... // must have the same number of partitions and cardinality as seriesX //皮尔逊相关系数:pearson //斯皮尔曼等级相关系数:spearman Double correlation = Statistics.corr(seriesX.srdd(), seriesY.srdd(), "pearson"); JavaRDD<Vector> data = ... // note that each Vector is a row and not a column // calculate the correlation matrix using Pearson's method. Use "spearman" for Spearman's method. // If a method is not specified, Pearson's method will be used by default. Matrix correlMatrix = Statistics.corr(data.rdd(), "pearson"); 3 分层抽样
JavaSparkContext jsc = ... JavaPairRDD<K, V> data = ... // an RDD of any key value pairs Map<K, Object> fractions = ... // specify the exact fraction desired from each key // Get an exact sample from each stratum JavaPairRDD<K, V> approxSample = data.sampleByKey(false, fractions); JavaPairRDD<K, V> exactSample = data.sampleByKeyExact(false, fractions); 4 假设检定
皮尔森卡方检定
JavaSparkContext jsc = ... Vector vec = ... // a vector composed of the frequencies of events // compute the goodness of fit. If a second vector to test against is not supplied as a parameter, // the test runs against a uniform distribution. ChiSqTestResult goodnessOfFitTestResult = Statistics.chiSqTest(vec); // summary of the test including the p-value, degrees of freedom, test statistic, the method used, // and the null hypothesis. System.out.println(goodnessOfFitTestResult); Matrix mat = ... // a contingency matrix // conduct Pearson's independence test on the input contingency matrix ChiSqTestResult independenceTestResult = Statistics.chiSqTest(mat); // summary of the test including the p-value, degrees of freedom... System.out.println(independenceTestResult); JavaRDD<LabeledPoint> obs = ... // an RDD of labeled points // The contingency table is constructed from the raw (feature, label) pairs and used to conduct // the independence test. Returns an array containing the ChiSquaredTestResult for every feature // against the label. ChiSqTestResult[] featureTestResults = Statistics.chiSqTest(obs.rdd()); int i = 1; for (ChiSqTestResult result : featureTestResults) { System.out.println("Column " + i + ":"); System.out.println(result); // summary of the test i++; } 1-sample, 2-sided Kolmogorov-Smirnov
JavaSparkContext jsc = ... JavaDoubleRDD data = jsc.parallelizeDoubles(Arrays.asList(0.2, 1.0, ...)); KolmogorovSmirnovTestResult testResult = Statistics.kolmogorovSmirnovTest(data, "norm", 0.0, 1.0); // summary of the test including the p-value, test statistic, // and null hypothesis // if our p-value indicates significance, we can reject the null hypothesis System.out.println(testResult); streaming significance testing
5 随机数生成
JavaSparkContext jsc = ... //均匀分布 uniform //标准正态分布 standard normal //泊松分布 Poisson JavaDoubleRDD u = normalJavaRDD(jsc, 1000000L, 10); JavaDoubleRDD v = u.map( new Function<Double, Double>() { public Double call(Double x) { return 1.0 + 2.0 * x; } }); 6 核密度估计
RDD<Double> data = ... // an RDD of sample data // Construct the density estimator with the sample data and a standard deviation for the Gaussian // kernels KernelDensity kd = new KernelDensity() .setSample(data) .setBandwidth(3.0); // Find density estimates for the given values double[] densities = kd.estimate(new double[] {-1.0, 2.0, 5.0}); 1.3 分类与回归
| 问题类型 | 支持的方法 |
|---|---|
| 二分类 | 线性支持向量机、逻辑回归、决策树、随即森林、梯度提升树、朴素贝叶斯 |
| 多分类 | 逻辑回归、决策树、随即森林、朴素贝叶斯 |
| 回归 | 线性最小二乘、Lasso、岭回归、决策树、随即森林、梯度提升树、保序回归 |
1 线性模型
- SVMWithSGD
- LogisticRegressionWithLBFGS
- LogisticRegressionWithSGD
- LinearRegressionWithSGD
- RidgeRegressionWithSGD
- LassoWithSGD
数学公式
目标函数包含两部分:正规化(regularizer)和损失函数。
正规化用来控制模型的复杂度,损失用来度量模型在训练中的错误。
损失函数:
- 合页损失(hinge loss)
- 逻辑损失(logistic loss)
- 平方损失(squared loss)
正规化:
- L2
- L1
- elastic net
最优化:
- SGD(Stochastic Gradient Descent-随机梯度下降)
- L-BFGS(Limited-Memory Broyden–Fletcher–Goldfarb–Shanno)
分类
线性支持向量机
public class SVMClassifier { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("SVM Classifier Example"); SparkContext sc = new SparkContext(conf); String path = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint> training = data.sample(false, 0.6, 11L); training.cache(); JavaRDD<LabeledPoint> test = data.subtract(training); // Run training algorithm to build the model. int numIterations = 100; final SVMModel model = SVMWithSGD.train(training.rdd(), numIterations); SVMWithSGD svmAlg = new SVMWithSGD(); svmAlg.optimizer() .setNumIterations(200) .setRegParam(0.1) .setUpdater(new L1Updater()); final SVMModel modelL1 = svmAlg.run(training.rdd()); // Clear the default threshold. model.clearThreshold(); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> scoreAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double score = model.predict(p.features()); return new Tuple2<Object, Object>(score, p.label()); } } ); // Get evaluation metrics. BinaryClassificationMetrics metrics = new BinaryClassificationMetrics(JavaRDD.toRDD(scoreAndLabels)); double auROC = metrics.areaUnderROC(); System.out.println("Area under ROC = " + auROC); // Save and load model model.save(sc, "myModelPath"); SVMModel sameModel = SVMModel.load(sc, "myModelPath"); } } 逻辑回归
public class MultinomialLogisticRegressionExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("LogisticRegression Classifier Example"); SparkContext sc = new SparkContext(conf); String path = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[] {0.6, 0.4}, 11L); JavaRDD<LabeledPoint> training = splits[0].cache(); JavaRDD<LabeledPoint> test = splits[1]; // Run training algorithm to build the model. final LogisticRegressionModel model = new LogisticRegressionWithLBFGS() .setNumClasses(10) .run(training.rdd()); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> predictionAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double prediction = model.predict(p.features()); return new Tuple2<Object, Object>(prediction, p.label()); } } ); // Get evaluation metrics. MulticlassMetrics metrics = new MulticlassMetrics(predictionAndLabels.rdd()); double precision = metrics.precision(); System.out.println("Precision = " + precision); // Save and load model model.save(sc, "myModelPath"); LogisticRegressionModel sameModel = LogisticRegressionModel.load(sc, "myModelPath"); } } 回归
线性最小二乘、Lasso、岭回归
public class LinearRegression { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("Linear Regression Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse the data String path = "data/mllib/ridge-data/lpsa.data"; JavaRDD<String> data = sc.textFile(path); JavaRDD<LabeledPoint> parsedData = data.map( new Function<String, LabeledPoint>() { public LabeledPoint call(String line) { String[] parts = line.split(","); String[] features = parts[1].split(" "); double[] v = new double[features.length]; for (int i = 0; i < features.length - 1; i++) v[i] = Double.parseDouble(features[i]); return new LabeledPoint(Double.parseDouble(parts[0]), Vectors.dense(v)); } } ); parsedData.cache(); // Building the model int numIterations = 100; final LinearRegressionModel model = LinearRegressionWithSGD.train(JavaRDD.toRDD(parsedData), numIterations); // Evaluate model on training examples and compute training error JavaRDD<Tuple2<Double, Double>> valuesAndPreds = parsedData.map( new Function<LabeledPoint, Tuple2<Double, Double>>() { public Tuple2<Double, Double> call(LabeledPoint point) { double prediction = model.predict(point.features()); return new Tuple2<Double, Double>(prediction, point.label()); } } ); double MSE = new JavaDoubleRDD(valuesAndPreds.map( new Function<Tuple2<Double, Double>, Object>() { public Object call(Tuple2<Double, Double> pair) { return Math.pow(pair._1() - pair._2(), 2.0); } } ).rdd()).mean(); System.out.println("training Mean Squared Error = " + MSE); // Save and load model model.save(sc.sc(), "myModelPath"); LinearRegressionModel sameModel = LinearRegressionModel.load(sc.sc(), "myModelPath"); } } 2 决策树
节点不纯和信息增益
- 节点不纯用来度量节点上标签的同质,实现包括分类模型中的基尼不纯和熵(Gini impurity and entropy)、回归模型中的方差(variance)。
- 信息增益用来度量父节点不纯与两个子节点不纯的加权和的差异。
停止规则
- 最大树深度maxDepth
- 最小信息增益minInfoGain
- 最小子节点实例数minInstancesPerNode
分类
examples/src/main/java/org/apache/spark/examples/mllib/JavaDecisionTreeClassificationExample.java
SparkConf sparkConf = new SparkConf().setAppName("JavaDecisionTreeClassificationExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Integer numClasses = 2; Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); String impurity = "gini"; Integer maxDepth = 5; Integer maxBins = 32; // Train a DecisionTree model for classification. final DecisionTreeModel model = DecisionTree.trainClassifier(trainingData, numClasses, categoricalFeaturesInfo, impurity, maxDepth, maxBins); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { @Override public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification tree model:/n" + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myDecisionTreeClassificationModel"); DecisionTreeModel sameModel = DecisionTreeModel .load(jsc.sc(), "target/tmp/myDecisionTreeClassificationModel"); 回归
examples/src/main/java/org/apache/spark/examples/mllib/JavaDecisionTreeRegressionExample.java
SparkConf sparkConf = new SparkConf().setAppName("JavaDecisionTreeRegressionExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); String impurity = "variance"; Integer maxDepth = 5; Integer maxBins = 32; // Train a DecisionTree model. final DecisionTreeModel model = DecisionTree.trainRegressor(trainingData, categoricalFeaturesInfo, impurity, maxDepth, maxBins); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testMSE = predictionAndLabel.map(new Function<Tuple2<Double, Double>, Double>() { @Override public Double call(Tuple2<Double, Double> pl) { Double diff = pl._1() - pl._2(); return diff * diff; } }).reduce(new Function2<Double, Double, Double>() { @Override public Double call(Double a, Double b) { return a + b; } }) / data.count(); System.out.println("Test Mean Squared Error: " + testMSE); System.out.println("Learned regression tree model:/n" + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myDecisionTreeRegressionModel"); DecisionTreeModel sameModel = DecisionTreeModel .load(jsc.sc(), "target/tmp/myDecisionTreeRegressionModel"); 3 集成树
随机森林和梯度提升树(Random Forests and Gradient-Boosted Trees)
- GradientBoostedTrees
- RandomForest
随机森林
分类
examples/src/main/java/org/apache/spark/examples/mllib/JavaRandomForestClassificationExample.java
SparkConf sparkConf = new SparkConf().setAppName("JavaRandomForestClassificationExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Train a RandomForest model. // Empty categoricalFeaturesInfo indicates all features are continuous. Integer numClasses = 2; HashMap<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); Integer numTrees = 3; // Use more in practice. String featureSubsetStrategy = "auto"; // Let the algorithm choose. String impurity = "gini"; Integer maxDepth = 5; Integer maxBins = 32; Integer seed = 12345; final RandomForestModel model = RandomForest.trainClassifier(trainingData, numClasses, categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins, seed); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { @Override public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification forest model:/n" + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myRandomForestClassificationModel"); RandomForestModel sameModel = RandomForestModel.load(jsc.sc(), "target/tmp/myRandomForestClassificationModel"); 回归
examples/src/main/java/org/apache/spark/examples/mllib/JavaRandomForestRegressionExample.java
SparkConf sparkConf = new SparkConf().setAppName("JavaRandomForestRegressionExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); Integer numTrees = 3; // Use more in practice. String featureSubsetStrategy = "auto"; // Let the algorithm choose. String impurity = "variance"; Integer maxDepth = 4; Integer maxBins = 32; Integer seed = 12345; // Train a RandomForest model. final RandomForestModel model = RandomForest.trainRegressor(trainingData, categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins, seed); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testMSE = predictionAndLabel.map(new Function<Tuple2<Double, Double>, Double>() { @Override public Double call(Tuple2<Double, Double> pl) { Double diff = pl._1() - pl._2(); return diff * diff; } }).reduce(new Function2<Double, Double, Double>() { @Override public Double call(Double a, Double b) { return a + b; } }) / testData.count(); System.out.println("Test Mean Squared Error: " + testMSE); System.out.println("Learned regression forest model:/n" + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myRandomForestRegressionModel"); RandomForestModel sameModel = RandomForestModel.load(jsc.sc(), "target/tmp/myRandomForestRegressionModel"); 梯度提升树
分类
examples/src/main/java/org/apache/spark/examples/mllib/JavaGradientBoostingClassificationExample.java
SparkConf sparkConf = new SparkConf() .setAppName("JavaGradientBoostedTreesClassificationExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Train a GradientBoostedTrees model. // The defaultParams for Classification use LogLoss by default. BoostingStrategy boostingStrategy = BoostingStrategy.defaultParams("Classification"); boostingStrategy.setNumIterations(3); // Note: Use more iterations in practice. boostingStrategy.getTreeStrategy().setNumClasses(2); boostingStrategy.getTreeStrategy().setMaxDepth(5); // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); boostingStrategy.treeStrategy().setCategoricalFeaturesInfo(categoricalFeaturesInfo); final GradientBoostedTreesModel model = GradientBoostedTrees.train(trainingData, boostingStrategy); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { @Override public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification GBT model:/n" + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myGradientBoostingClassificationModel"); GradientBoostedTreesModel sameModel = GradientBoostedTreesModel.load(jsc.sc(), "target/tmp/myGradientBoostingClassificationModel"); 回归
examples/src/main/java/org/apache/spark/examples/mllib/JavaGradientBoostingRegressionExample.java
SparkConf sparkConf = new SparkConf() .setAppName("JavaGradientBoostedTreesRegressionExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Train a GradientBoostedTrees model. // The defaultParams for Regression use SquaredError by default. BoostingStrategy boostingStrategy = BoostingStrategy.defaultParams("Regression"); boostingStrategy.setNumIterations(3); // Note: Use more iterations in practice. boostingStrategy.getTreeStrategy().setMaxDepth(5); // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); boostingStrategy.treeStrategy().setCategoricalFeaturesInfo(categoricalFeaturesInfo); final GradientBoostedTreesModel model = GradientBoostedTrees.train(trainingData, boostingStrategy); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testMSE = predictionAndLabel.map(new Function<Tuple2<Double, Double>, Double>() { @Override public Double call(Tuple2<Double, Double> pl) { Double diff = pl._1() - pl._2(); return diff * diff; } }).reduce(new Function2<Double, Double, Double>() { @Override public Double call(Double a, Double b) { return a + b; } }) / data.count(); System.out.println("Test Mean Squared Error: " + testMSE); System.out.println("Learned regression GBT model:/n" + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myGradientBoostingRegressionModel"); GradientBoostedTreesModel sameModel = GradientBoostedTreesModel.load(jsc.sc(), "target/tmp/myGradientBoostingRegressionModel"); 4 朴素贝叶斯
- 多项式模型 以单词为粒度 “multinomial”
- 伯努利模型 以文件为粒度 “bernoulli”
examples/src/main/java/org/apache/spark/examples/mllib/JavaNaiveBayesExample.java
String path = "data/mllib/sample_naive_bayes_data.txt"; JavaRDD<LabeledPoint> inputData = MLUtils.loadLibSVMFile(jsc.sc(), path).toJavaRDD(); JavaRDD<LabeledPoint>[] tmp = inputData.randomSplit(new double[]{0.6, 0.4}, 12345); JavaRDD<LabeledPoint> training = tmp[0]; // training set JavaRDD<LabeledPoint> test = tmp[1]; // test set final NaiveBayesModel model = NaiveBayes.train(training.rdd(), 1.0); JavaPairRDD<Double, Double> predictionAndLabel = test.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); double accuracy = predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { @Override public Boolean call(Tuple2<Double, Double> pl) { return pl._1().equals(pl._2()); } }).count() / (double) test.count(); // Save and load model model.save(jsc.sc(), "target/tmp/myNaiveBayesModel"); NaiveBayesModel sameModel = NaiveBayesModel.load(jsc.sc(), "target/tmp/myNaiveBayesModel"); 5 保序回归
examples/src/main/java/org/apache/spark/examples/mllib/JavaIsotonicRegressionExample.java
JavaRDD<String> data = jsc.textFile("data/mllib/sample_isotonic_regression_data.txt"); // Create label, feature, weight tuples from input data with weight set to default value 1.0. JavaRDD<Tuple3<Double, Double, Double>> parsedData = data.map( new Function<String, Tuple3<Double, Double, Double>>() { public Tuple3<Double, Double, Double> call(String line) { String[] parts = line.split(","); return new Tuple3<>(new Double(parts[0]), new Double(parts[1]), 1.0); } } ); // Split data into training (60%) and test (40%) sets. JavaRDD<Tuple3<Double, Double, Double>>[] splits = parsedData.randomSplit(new double[]{0.6, 0.4}, 11L); JavaRDD<Tuple3<Double, Double, Double>> training = splits[0]; JavaRDD<Tuple3<Double, Double, Double>> test = splits[1]; // Create isotonic regression model from training data. // Isotonic parameter defaults to true so it is only shown for demonstration final IsotonicRegressionModel model = new IsotonicRegression().setIsotonic(true).run(training); // Create tuples of predicted and real labels. JavaPairRDD<Double, Double> predictionAndLabel = test.mapToPair( new PairFunction<Tuple3<Double, Double, Double>, Double, Double>() { @Override public Tuple2<Double, Double> call(Tuple3<Double, Double, Double> point) { Double predictedLabel = model.predict(point._2()); return new Tuple2<Double, Double>(predictedLabel, point._1()); } } ); // Calculate mean squared error between predicted and real labels. Double meanSquaredError = new JavaDoubleRDD(predictionAndLabel.map( new Function<Tuple2<Double, Double>, Object>() { @Override public Object call(Tuple2<Double, Double> pl) { return Math.pow(pl._1() - pl._2(), 2); } } ).rdd()).mean(); System.out.println("Mean Squared Error = " + meanSquaredError); // Save and load model model.save(jsc.sc(), "target/tmp/myIsotonicRegressionModel"); IsotonicRegressionModel sameModel = IsotonicRegressionModel.load(jsc.sc(), "target/tmp/myIsotonicRegressionModel"); 1.4 协同过滤
交替最小二乘(ALS)
显式反馈和隐式反馈
SparkConf conf = new SparkConf().setAppName("Java Collaborative Filtering Example"); JavaSparkContext jsc = new JavaSparkContext(conf); // Load and parse the data String path = "data/mllib/als/test.data"; JavaRDD<String> data = jsc.textFile(path); JavaRDD<Rating> ratings = data.map( new Function<String, Rating>() { public Rating call(String s) { String[] sarray = s.split(","); return new Rating(Integer.parseInt(sarray[0]), Integer.parseInt(sarray[1]), Double.parseDouble(sarray[2])); } } ); // Build the recommendation model using ALS int rank = 10; int numIterations = 10; MatrixFactorizationModel model = ALS.train(JavaRDD.toRDD(ratings), rank, numIterations, 0.01); // Evaluate the model on rating data JavaRDD<Tuple2<Object, Object>> userProducts = ratings.map( new Function<Rating, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(Rating r) { return new Tuple2<Object, Object>(r.user(), r.product()); } } ); JavaPairRDD<Tuple2<Integer, Integer>, Double> predictions = JavaPairRDD.fromJavaRDD( model.predict(JavaRDD.toRDD(userProducts)).toJavaRDD().map( new Function<Rating, Tuple2<Tuple2<Integer, Integer>, Double>>() { public Tuple2<Tuple2<Integer, Integer>, Double> call(Rating r){ return new Tuple2<Tuple2<Integer, Integer>, Double>( new Tuple2<Integer, Integer>(r.user(), r.product()), r.rating()); } } )); JavaRDD<Tuple2<Double, Double>> ratesAndPreds = JavaPairRDD.fromJavaRDD(ratings.map( new Function<Rating, Tuple2<Tuple2<Integer, Integer>, Double>>() { public Tuple2<Tuple2<Integer, Integer>, Double> call(Rating r){ return new Tuple2<Tuple2<Integer, Integer>, Double>( new Tuple2<Integer, Integer>(r.user(), r.product()), r.rating()); } } )).join(predictions).values(); double MSE = JavaDoubleRDD.fromRDD(ratesAndPreds.map( new Function<Tuple2<Double, Double>, Object>() { public Object call(Tuple2<Double, Double> pair) { Double err = pair._1() - pair._2(); return err * err; } } ).rdd()).mean(); System.out.println("Mean Squared Error = " + MSE); // Save and load model model.save(jsc.sc(), "target/tmp/myCollaborativeFilter"); MatrixFactorizationModel sameModel = MatrixFactorizationModel.load(jsc.sc(), "target/tmp/myCollaborativeFilter"); 1.5 聚类
1 K均值
public class KMeansExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("K-means Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse data String path = "data/mllib/kmeans_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) values[i] = Double.parseDouble(sarray[i]); return Vectors.dense(values); } } ); parsedData.cache(); // Cluster the data into two classes using KMeans int numClusters = 2; int numIterations = 20; KMeansModel clusters = KMeans.train(parsedData.rdd(), numClusters, numIterations); // Evaluate clustering by computing Within Set Sum of Squared Errors double WSSSE = clusters.computeCost(parsedData.rdd()); System.out.println("Within Set Sum of Squared Errors = " + WSSSE); // Save and load model clusters.save(sc.sc(), "myModelPath"); KMeansModel sameModel = KMeansModel.load(sc.sc(), "myModelPath"); } } 2 高斯混合
public class GaussianMixtureExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("GaussianMixture Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse data String path = "data/mllib/gmm_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.trim().split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) values[i] = Double.parseDouble(sarray[i]); return Vectors.dense(values); } } ); parsedData.cache(); // Cluster the data into two classes using GaussianMixture GaussianMixtureModel gmm = new GaussianMixture().setK(2).run(parsedData.rdd()); // Save and load GaussianMixtureModel gmm.save(sc.sc(), "myGMMModel"); GaussianMixtureModel sameModel = GaussianMixtureModel.load(sc.sc(), "myGMMModel"); // Output the parameters of the mixture model for(int j=0; j<gmm.k(); j++) { System.out.printf("weight=%f/nmu=%s/nsigma=/n%s/n", gmm.weights()[j], gmm.gaussians()[j].mu(), gmm.gaussians()[j].sigma()); } } } 3 幂迭代聚类(PIC)
// Load and parse the data JavaRDD<String> data = sc.textFile("data/mllib/pic_data.txt"); JavaRDD<Tuple3<Long, Long, Double>> similarities = data.map( new Function<String, Tuple3<Long, Long, Double>>() { public Tuple3<Long, Long, Double> call(String line) { String[] parts = line.split(" "); return new Tuple3<>(new Long(parts[0]), new Long(parts[1]), new Double(parts[2])); } } ); // Cluster the data into two classes using PowerIterationClustering PowerIterationClustering pic = new PowerIterationClustering() .setK(2) .setMaxIterations(10); PowerIterationClusteringModel model = pic.run(similarities); for (PowerIterationClustering.Assignment a: model.assignments().toJavaRDD().collect()) { System.out.println(a.id() + " -> " + a.cluster()); } // Save and load model model.save(sc.sc(), "myModelPath"); PowerIterationClusteringModel sameModel = PowerIterationClusteringModel.load(sc.sc(), "myModelPath"); 4 LDA
public class JavaLDAExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("LDA Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse the data String path = "data/mllib/sample_lda_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.trim().split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) values[i] = Double.parseDouble(sarray[i]); return Vectors.dense(values); } } ); // Index documents with unique IDs JavaPairRDD<Long, Vector> corpus = JavaPairRDD.fromJavaRDD(parsedData.zipWithIndex().map( new Function<Tuple2<Vector, Long>, Tuple2<Long, Vector>>() { public Tuple2<Long, Vector> call(Tuple2<Vector, Long> doc_id) { return doc_id.swap(); } } )); corpus.cache(); // Cluster the documents into three topics using LDA DistributedLDAModel ldaModel = new LDA().setK(3).run(corpus); // Output topics. Each is a distribution over words (matching word count vectors) System.out.println("Learned topics (as distributions over vocab of " + ldaModel.vocabSize() + " words):"); Matrix topics = ldaModel.topicsMatrix(); for (int topic = 0; topic < 3; topic++) { System.out.print("Topic " + topic + ":"); for (int word = 0; word < ldaModel.vocabSize(); word++) { System.out.print(" " + topics.apply(word, topic)); } System.out.println(); } ldaModel.save(sc.sc(), "myLDAModel"); DistributedLDAModel sameModel = DistributedLDAModel.load(sc.sc(), "myLDAModel"); } } 5 二分K均值
ArrayList<Vector> localData = Lists.newArrayList( Vectors.dense(0.1, 0.1), Vectors.dense(0.3, 0.3), Vectors.dense(10.1, 10.1), Vectors.dense(10.3, 10.3), Vectors.dense(20.1, 20.1), Vectors.dense(20.3, 20.3), Vectors.dense(30.1, 30.1), Vectors.dense(30.3, 30.3) ); JavaRDD<Vector> data = sc.parallelize(localData, 2); BisectingKMeans bkm = new BisectingKMeans() .setK(4); BisectingKMeansModel model = bkm.run(data); System.out.println("Compute Cost: " + model.computeCost(data)); for (Vector center: model.clusterCenters()) { System.out.println(""); } Vector[] clusterCenters = model.clusterCenters(); for (int i = 0; i < clusterCenters.length; i++) { Vector clusterCenter = clusterCenters[i]; System.out.println("Cluster Center " + i + ": " + clusterCenter); } 6 流式K均值
val trainingData = ssc.textFileStream("/training/data/dir").map(Vectors.parse) val testData = ssc.textFileStream("/testing/data/dir").map(LabeledPoint.parse) val numDimensions = 3 val numClusters = 2 val model = new StreamingKMeans() .setK(numClusters) .setDecayFactor(1.0) .setRandomCenters(numDimensions, 0.0) model.trainOn(trainingData) model.predictOnValues(testData.map(lp => (lp.label, lp.features))).print() ssc.start() ssc.awaitTermination() 1.6 降维
1 奇异值分解(SVD)
public class SVD { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("SVD Example"); SparkContext sc = new SparkContext(conf); double[][] array = ... LinkedList<Vector> rowsList = new LinkedList<Vector>(); for (int i = 0; i < array.length; i++) { Vector currentRow = Vectors.dense(array[i]); rowsList.add(currentRow); } JavaRDD<Vector> rows = JavaSparkContext.fromSparkContext(sc).parallelize(rowsList); // Create a RowMatrix from JavaRDD<Vector>. RowMatrix mat = new RowMatrix(rows.rdd()); // Compute the top 4 singular values and corresponding singular vectors. SingularValueDecomposition<RowMatrix, Matrix> svd = mat.computeSVD(4, true, 1.0E-9d); RowMatrix U = svd.U(); Vector s = svd.s(); Matrix V = svd.V(); } } 2 主成分分析(PCA)
public class PCA { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("PCA Example"); SparkContext sc = new SparkContext(conf); double[][] array = ... LinkedList<Vector> rowsList = new LinkedList<Vector>(); for (int i = 0; i < array.length; i++) { Vector currentRow = Vectors.dense(array[i]); rowsList.add(currentRow); } JavaRDD<Vector> rows = JavaSparkContext.fromSparkContext(sc).parallelize(rowsList); // Create a RowMatrix from JavaRDD<Vector>. RowMatrix mat = new RowMatrix(rows.rdd()); // Compute the top 3 principal components. Matrix pc = mat.computePrincipalComponents(3); RowMatrix projected = mat.multiply(pc); } } 1.7 特征提取和转换
TF-IDF
val sc: SparkContext = ... // Load documents (one per line). val documents: RDD[Seq[String]] = sc.textFile("...").map(_.split(" ").toSeq) val hashingTF = new HashingTF() val tf: RDD[Vector] = hashingTF.transform(documents) tf.cache() val idf = new IDF().fit(tf) val tfidf: RDD[Vector] = idf.transform(tf) tf.cache() val idf = new IDF(minDocFreq = 2).fit(tf) val tfidf: RDD[Vector] = idf.transform(tf) Word2Vec
val input = sc.textFile("text8").map(line => line.split(" ").toSeq) val word2vec = new Word2Vec() val model = word2vec.fit(input) val synonyms = model.findSynonyms("china", 40) for((synonym, cosineSimilarity) <- synonyms) { println(s"$synonym $cosineSimilarity") } // Save and load model model.save(sc, "myModelPath") val sameModel = Word2VecModel.load(sc, "myModelPath") 标准化(StandardScaler)
val data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") val scaler1 = new StandardScaler().fit(data.map(x => x.features)) val scaler2 = new StandardScaler(withMean = true, withStd = true).fit(data.map(x => x.features)) // scaler3 is an identical model to scaler2, and will produce identical transformations val scaler3 = new StandardScalerModel(scaler2.std, scaler2.mean) // data1 will be unit variance. val data1 = data.map(x => (x.label, scaler1.transform(x.features))) // Without converting the features into dense vectors, transformation with zero mean will raise // exception on sparse vector. // data2 will be unit variance and zero mean. val data2 = data.map(x => (x.label, scaler2.transform(Vectors.dense(x.features.toArray)))) 归一化(Normalizer)
val data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") val normalizer1 = new Normalizer() val normalizer2 = new Normalizer(p = Double.PositiveInfinity) // Each sample in data1 will be normalized using $L^2$ norm. val data1 = data.map(x => (x.label, normalizer1.transform(x.features))) // Each sample in data2 will be normalized using $L^/infty$ norm. val data2 = data.map(x => (x.label, normalizer2.transform(x.features))) 卡方选择(ChiSqSelector)
SparkConf sparkConf = new SparkConf().setAppName("JavaChiSqSelector"); JavaSparkContext sc = new JavaSparkContext(sparkConf); JavaRDD<LabeledPoint> points = MLUtils.loadLibSVMFile(sc.sc(), "data/mllib/sample_libsvm_data.txt").toJavaRDD().cache(); // Discretize data in 16 equal bins since ChiSqSelector requires categorical features // Even though features are doubles, the ChiSqSelector treats each unique value as a category JavaRDD<LabeledPoint> discretizedData = points.map( new Function<LabeledPoint, LabeledPoint>() { @Override public LabeledPoint call(LabeledPoint lp) { final double[] discretizedFeatures = new double[lp.features().size()]; for (int i = 0; i < lp.features().size(); ++i) { discretizedFeatures[i] = Math.floor(lp.features().apply(i) / 16); } return new LabeledPoint(lp.label(), Vectors.dense(discretizedFeatures)); } }); // Create ChiSqSelector that will select top 50 of 692 features ChiSqSelector selector = new ChiSqSelector(50); // Create ChiSqSelector model (selecting features) final ChiSqSelectorModel transformer = selector.fit(discretizedData.rdd()); // Filter the top 50 features from each feature vector JavaRDD<LabeledPoint> filteredData = discretizedData.map( new Function<LabeledPoint, LabeledPoint>() { @Override public LabeledPoint call(LabeledPoint lp) { return new LabeledPoint(lp.label(), transformer.transform(lp.features())); } } ); sc.stop(); ElementwiseProduct
// Create some vector data; also works for sparse vectors JavaRDD<Vector> data = sc.parallelize(Arrays.asList( Vectors.dense(1.0, 2.0, 3.0), Vectors.dense(4.0, 5.0, 6.0))); Vector transformingVector = Vectors.dense(0.0, 1.0, 2.0); ElementwiseProduct transformer = new ElementwiseProduct(transformingVector); // Batch transform and per-row transform give the same results: JavaRDD<Vector> transformedData = transformer.transform(data); JavaRDD<Vector> transformedData2 = data.map( new Function<Vector, Vector>() { @Override public Vector call(Vector v) { return transformer.transform(v); } } ); PCA
val data = sc.textFile("data/mllib/ridge-data/lpsa.data").map { line => val parts = line.split(',') LabeledPoint(parts(0).toDouble, Vectors.dense(parts(1).split(' ').map(_.toDouble))) }.cache() val splits = data.randomSplit(Array(0.6, 0.4), seed = 11L) val training = splits(0).cache() val test = splits(1) val pca = new PCA(training.first().features.size/2).fit(data.map(_.features)) val training_pca = training.map(p => p.copy(features = pca.transform(p.features))) val test_pca = test.map(p => p.copy(features = pca.transform(p.features))) val numIterations = 100 val model = LinearRegressionWithSGD.train(training, numIterations) val model_pca = LinearRegressionWithSGD.train(training_pca, numIterations) val valuesAndPreds = test.map { point => val score = model.predict(point.features) (score, point.label) } val valuesAndPreds_pca = test_pca.map { point => val score = model_pca.predict(point.features) (score, point.label) } val MSE = valuesAndPreds.map{case(v, p) => math.pow((v - p), 2)}.mean() val MSE_pca = valuesAndPreds_pca.map{case(v, p) => math.pow((v - p), 2)}.mean() println("Mean Squared Error = " + MSE) println("PCA Mean Squared Error = " + MSE_pca) 1.8 频繁模式挖掘(FPM)
FP-growth
examples/src/main/java/org/apache/spark/examples/mllib/JavaSimpleFPGrowth.java
JavaRDD<String> data = sc.textFile("data/mllib/sample_fpgrowth.txt"); JavaRDD<List<String>> transactions = data.map( new Function<String, List<String>>() { public List<String> call(String line) { String[] parts = line.split(" "); return Arrays.asList(parts); } } ); FPGrowth fpg = new FPGrowth() .setMinSupport(0.2) .setNumPartitions(10); FPGrowthModel<String> model = fpg.run(transactions); for (FPGrowth.FreqItemset<String> itemset: model.freqItemsets().toJavaRDD().collect()) { System.out.println("[" + itemset.javaItems() + "], " + itemset.freq()); } double minConfidence = 0.8; for (AssociationRules.Rule<String> rule : model.generateAssociationRules(minConfidence).toJavaRDD().collect()) { System.out.println( rule.javaAntecedent() + " => " + rule.javaConsequent() + ", " + rule.confidence()); } Association Rules
examples/src/main/java/org/apache/spark/examples/mllib/JavaAssociationRulesExample.java
JavaRDD<FPGrowth.FreqItemset<String>> freqItemsets = sc.parallelize(Arrays.asList( new FreqItemset<String>(new String[] {"a"}, 15L), new FreqItemset<String>(new String[] {"b"}, 35L), new FreqItemset<String>(new String[] {"a", "b"}, 12L) )); AssociationRules arules = new AssociationRules() .setMinConfidence(0.8); JavaRDD<AssociationRules.Rule<String>> results = arules.run(freqItemsets); for (AssociationRules.Rule<String> rule : results.collect()) { System.out.println( rule.javaAntecedent() + " => " + rule.javaConsequent() + ", " + rule.confidence()); } PrefixSpan
examples/src/main/java/org/apache/spark/examples/mllib/JavaPrefixSpanExample.java
JavaRDD<List<List<Integer>>> sequences = sc.parallelize(Arrays.asList( Arrays.asList(Arrays.asList(1, 2), Arrays.asList(3)), Arrays.asList(Arrays.asList(1), Arrays.asList(3, 2), Arrays.asList(1, 2)), Arrays.asList(Arrays.asList(1, 2), Arrays.asList(5)), Arrays.asList(Arrays.asList(6)) ), 2); PrefixSpan prefixSpan = new PrefixSpan() .setMinSupport(0.5) .setMaxPatternLength(5); PrefixSpanModel<Integer> model = prefixSpan.run(sequences); for (PrefixSpan.FreqSequence<Integer> freqSeq: model.freqSequences().toJavaRDD().collect()) { System.out.println(freqSeq.javaSequence() + ", " + freqSeq.freq()); } 1.9 评估指标
分类模型评估
- True Positive (TP) - label is positive and prediction is also positive
- True Negative (TN) - label is negative and prediction is also negative
- False Positive (FP) - label is negative but prediction is positive
- False Negative (FN) - label is positive but prediction is negative
二分类
- Precision (Postive Predictive Value)
- Recall (True Positive Rate)
- F-measure
- Receiver Operating Characteristic (ROC)
- Area Under ROC Curve
- Area Under Precision-Recall Curve
examples/src/main/java/org/apache/spark/examples/mllib/JavaBinaryClassificationMetricsExample.java
String path = "data/mllib/sample_binary_classification_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.6, 0.4}, 11L); JavaRDD<LabeledPoint> training = splits[0].cache(); JavaRDD<LabeledPoint> test = splits[1]; // Run training algorithm to build the model. final LogisticRegressionModel model = new LogisticRegressionWithLBFGS() .setNumClasses(2) .run(training.rdd()); // Clear the prediction threshold so the model will return probabilities model.clearThreshold(); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> predictionAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double prediction = model.predict(p.features()); return new Tuple2<Object, Object>(prediction, p.label()); } } ); // Get evaluation metrics. BinaryClassificationMetrics metrics = new BinaryClassificationMetrics(predictionAndLabels.rdd()); // Precision by threshold JavaRDD<Tuple2<Object, Object>> precision = metrics.precisionByThreshold().toJavaRDD(); System.out.println("Precision by threshold: " + precision.toArray()); // Recall by threshold JavaRDD<Tuple2<Object, Object>> recall = metrics.recallByThreshold().toJavaRDD(); System.out.println("Recall by threshold: " + recall.toArray()); // F Score by threshold JavaRDD<Tuple2<Object, Object>> f1Score = metrics.fMeasureByThreshold().toJavaRDD(); System.out.println("F1 Score by threshold: " + f1Score.toArray()); JavaRDD<Tuple2<Object, Object>> f2Score = metrics.fMeasureByThreshold(2.0).toJavaRDD(); System.out.println("F2 Score by threshold: " + f2Score.toArray()); // Precision-recall curve JavaRDD<Tuple2<Object, Object>> prc = metrics.pr().toJavaRDD(); System.out.println("Precision-recall curve: " + prc.toArray()); // Thresholds JavaRDD<Double> thresholds = precision.map( new Function<Tuple2<Object, Object>, Double>() { public Double call(Tuple2<Object, Object> t) { return new Double(t._1().toString()); } } ); // ROC Curve JavaRDD<Tuple2<Object, Object>> roc = metrics.roc().toJavaRDD(); System.out.println("ROC curve: " + roc.toArray()); // AUPRC System.out.println("Area under precision-recall curve = " + metrics.areaUnderPR()); // AUROC System.out.println("Area under ROC = " + metrics.areaUnderROC()); // Save and load model model.save(sc, "target/tmp/LogisticRegressionModel"); LogisticRegressionModel sameModel = LogisticRegressionModel.load(sc, "target/tmp/LogisticRegressionModel"); 多分类
examples/src/main/java/org/apache/spark/examples/mllib/JavaMulticlassClassificationMetricsExample.java
String path = "data/mllib/sample_multiclass_classification_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.6, 0.4}, 11L); JavaRDD<LabeledPoint> training = splits[0].cache(); JavaRDD<LabeledPoint> test = splits[1]; // Run training algorithm to build the model. final LogisticRegressionModel model = new LogisticRegressionWithLBFGS() .setNumClasses(3) .run(training.rdd()); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> predictionAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double prediction = model.predict(p.features()); return new Tuple2<Object, Object>(prediction, p.label()); } } ); // Get evaluation metrics. MulticlassMetrics metrics = new MulticlassMetrics(predictionAndLabels.rdd()); // Confusion matrix Matrix confusion = metrics.confusionMatrix(); System.out.println("Confusion matrix: /n" + confusion); // Overall statistics System.out.println("Precision = " + metrics.precision()); System.out.println("Recall = " + metrics.recall()); System.out.println("F1 Score = " + metrics.fMeasure()); // Stats by labels for (int i = 0; i < metrics.labels().length; i++) { System.out.format("Class %f precision = %f/n", metrics.labels()[i],metrics.precision (metrics.labels()[i])); System.out.format("Class %f recall = %f/n", metrics.labels()[i], metrics.recall(metrics .labels()[i])); System.out.format("Class %f F1 score = %f/n", metrics.labels()[i], metrics.fMeasure (metrics.labels()[i])); } //Weighted stats System.out.format("Weighted precision = %f/n", metrics.weightedPrecision()); System.out.format("Weighted recall = %f/n", metrics.weightedRecall()); System.out.format("Weighted F1 score = %f/n", metrics.weightedFMeasure()); System.out.format("Weighted false positive rate = %f/n", metrics.weightedFalsePositiveRate()); // Save and load model model.save(sc, "target/tmp/LogisticRegressionModel"); LogisticRegressionModel sameModel = LogisticRegressionModel.load(sc, "target/tmp/LogisticRegressionModel"); 多标签分类
examples/src/main/java/org/apache/spark/examples/mllib/JavaMultiLabelClassificationMetricsExample.java
List<Tuple2<double[], double[]>> data = Arrays.asList( new Tuple2<double[], double[]>(new double[]{0.0, 1.0}, new double[]{0.0, 2.0}), new Tuple2<double[], double[]>(new double[]{0.0, 2.0}, new double[]{0.0, 1.0}), new Tuple2<double[], double[]>(new double[]{}, new double[]{0.0}), new Tuple2<double[], double[]>(new double[]{2.0}, new double[]{2.0}), new Tuple2<double[], double[]>(new double[]{2.0, 0.0}, new double[]{2.0, 0.0}), new Tuple2<double[], double[]>(new double[]{0.0, 1.0, 2.0}, new double[]{0.0, 1.0}), new Tuple2<double[], double[]>(new double[]{1.0}, new double[]{1.0, 2.0}) ); JavaRDD<Tuple2<double[], double[]>> scoreAndLabels = sc.parallelize(data); // Instantiate metrics object MultilabelMetrics metrics = new MultilabelMetrics(scoreAndLabels.rdd()); // Summary stats System.out.format("Recall = %f/n", metrics.recall()); System.out.format("Precision = %f/n", metrics.precision()); System.out.format("F1 measure = %f/n", metrics.f1Measure()); System.out.format("Accuracy = %f/n", metrics.accuracy()); // Stats by labels for (int i = 0; i < metrics.labels().length - 1; i++) { System.out.format("Class %1.1f precision = %f/n", metrics.labels()[i], metrics.precision (metrics.labels()[i])); System.out.format("Class %1.1f recall = %f/n", metrics.labels()[i], metrics.recall(metrics .labels()[i])); System.out.format("Class %1.1f F1 score = %f/n", metrics.labels()[i], metrics.f1Measure (metrics.labels()[i])); } // Micro stats System.out.format("Micro recall = %f/n", metrics.microRecall()); System.out.format("Micro precision = %f/n", metrics.microPrecision()); System.out.format("Micro F1 measure = %f/n", metrics.microF1Measure()); // Hamming loss System.out.format("Hamming loss = %f/n", metrics.hammingLoss()); // Subset accuracy System.out.format("Subset accuracy = %f/n", metrics.subsetAccuracy()); Ranking系统
examples/src/main/java/org/apache/spark/examples/mllib/JavaRankingMetricsExample.java
String path = "data/mllib/sample_movielens_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<Rating> ratings = data.map( new Function<String, Rating>() { public Rating call(String line) { String[] parts = line.split("::"); return new Rating(Integer.parseInt(parts[0]), Integer.parseInt(parts[1]), Double .parseDouble(parts[2]) - 2.5); } } ); ratings.cache(); // Train an ALS model final MatrixFactorizationModel model = ALS.train(JavaRDD.toRDD(ratings), 10, 10, 0.01); // Get top 10 recommendations for every user and scale ratings from 0 to 1 JavaRDD<Tuple2<Object, Rating[]>> userRecs = model.recommendProductsForUsers(10).toJavaRDD(); JavaRDD<Tuple2<Object, Rating[]>> userRecsScaled = userRecs.map( new Function<Tuple2<Object, Rating[]>, Tuple2<Object, Rating[]>>() { public Tuple2<Object, Rating[]> call(Tuple2<Object, Rating[]> t) { Rating[] scaledRatings = new Rating[t._2().length]; for (int i = 0; i < scaledRatings.length; i++) { double newRating = Math.max(Math.min(t._2()[i].rating(), 1.0), 0.0); scaledRatings[i] = new Rating(t._2()[i].user(), t._2()[i].product(), newRating); } return new Tuple2<Object, Rating[]>(t._1(), scaledRatings); } } ); JavaPairRDD<Object, Rating[]> userRecommended = JavaPairRDD.fromJavaRDD(userRecsScaled); // Map ratings to 1 or 0, 1 indicating a movie that should be recommended JavaRDD<Rating> binarizedRatings = ratings.map( new Function<Rating, Rating>() { public Rating call(Rating r) { double binaryRating; if (r.rating() > 0.0) { binaryRating = 1.0; } else { binaryRating = 0.0; } return new Rating(r.user(), r.product(), binaryRating); } } ); // Group ratings by common user JavaPairRDD<Object, Iterable<Rating>> userMovies = binarizedRatings.groupBy( new Function<Rating, Object>() { public Object call(Rating r) { return r.user(); } } ); // Get true relevant documents from all user ratings JavaPairRDD<Object, List<Integer>> userMoviesList = userMovies.mapValues( new Function<Iterable<Rating>, List<Integer>>() { public List<Integer> call(Iterable<Rating> docs) { List<Integer> products = new ArrayList<Integer>(); for (Rating r : docs) { if (r.rating() > 0.0) { products.add(r.product()); } } return products; } } ); // Extract the product id from each recommendation JavaPairRDD<Object, List<Integer>> userRecommendedList = userRecommended.mapValues( new Function<Rating[], List<Integer>>() { public List<Integer> call(Rating[] docs) { List<Integer> products = new ArrayList<Integer>(); for (Rating r : docs) { products.add(r.product()); } return products; } } ); JavaRDD<Tuple2<List<Integer>, List<Integer>>> relevantDocs = userMoviesList.join (userRecommendedList).values(); // Instantiate the metrics object RankingMetrics metrics = RankingMetrics.of(relevantDocs); // Precision and NDCG at k Integer[] kVector = {1, 3, 5}; for (Integer k : kVector) { System.out.format("Precision at %d = %f/n", k, metrics.precisionAt(k)); System.out.format("NDCG at %d = %f/n", k, metrics.ndcgAt(k)); } // Mean average precision System.out.format("Mean average precision = %f/n", metrics.meanAveragePrecision()); // Evaluate the model using numerical ratings and regression metrics JavaRDD<Tuple2<Object, Object>> userProducts = ratings.map( new Function<Rating, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(Rating r) { return new Tuple2<Object, Object>(r.user(), r.product()); } } ); JavaPairRDD<Tuple2<Integer, Integer>, Object> predictions = JavaPairRDD.fromJavaRDD( model.predict(JavaRDD.toRDD(userProducts)).toJavaRDD().map( new Function<Rating, Tuple2<Tuple2<Integer, Integer>, Object>>() { public Tuple2<Tuple2<Integer, Integer>, Object> call(Rating r) { return new Tuple2<Tuple2<Integer, Integer>, Object>( new Tuple2<Integer, Integer>(r.user(), r.product()), r.rating()); } } )); JavaRDD<Tuple2<Object, Object>> ratesAndPreds = JavaPairRDD.fromJavaRDD(ratings.map( new Function<Rating, Tuple2<Tuple2<Integer, Integer>, Object>>() { public Tuple2<Tuple2<Integer, Integer>, Object> call(Rating r) { return new Tuple2<Tuple2<Integer, Integer>, Object>( new Tuple2<Integer, Integer>(r.user(), r.product()), r.rating()); } } )).join(predictions).values(); // Create regression metrics object RegressionMetrics regressionMetrics = new RegressionMetrics(ratesAndPreds.rdd()); // Root mean squared error System.out.format("RMSE = %f/n", regressionMetrics.rootMeanSquaredError()); // R-squared System.out.format("R-squared = %f/n", regressionMetrics.r2()); 回归模型评估
- Mean Squared Error (MSE)
- Root Mean Squared Error (RMSE)
- Mean Absoloute Error (MAE)
- Coefficient of Determination (R2)
examples/src/main/java/org/apache/spark/examples/mllib/JavaRegressionMetricsExample.java
// Load and parse the data String path = "data/mllib/sample_linear_regression_data.txt"; JavaRDD<String> data = sc.textFile(path); JavaRDD<LabeledPoint> parsedData = data.map( new Function<String, LabeledPoint>() { public LabeledPoint call(String line) { String[] parts = line.split(" "); double[] v = new double[parts.length - 1]; for (int i = 1; i < parts.length - 1; i++) v[i - 1] = Double.parseDouble(parts[i].split(":")[1]); return new LabeledPoint(Double.parseDouble(parts[0]), Vectors.dense(v)); } } ); parsedData.cache(); // Building the model int numIterations = 100; final LinearRegressionModel model = LinearRegressionWithSGD.train(JavaRDD.toRDD(parsedData), numIterations); // Evaluate model on training examples and compute training error JavaRDD<Tuple2<Object, Object>> valuesAndPreds = parsedData.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint point) { double prediction = model.predict(point.features()); return new Tuple2<Object, Object>(prediction, point.label()); } } ); // Instantiate metrics object RegressionMetrics metrics = new RegressionMetrics(valuesAndPreds.rdd()); // Squared error System.out.format("MSE = %f/n", metrics.meanSquaredError()); System.out.format("RMSE = %f/n", metrics.rootMeanSquaredError()); // R-squared System.out.format("R Squared = %f/n", metrics.r2()); // Mean absolute error System.out.format("MAE = %f/n", metrics.meanAbsoluteError()); // Explained variance System.out.format("Explained Variance = %f/n", metrics.explainedVariance()); // Save and load model model.save(sc.sc(), "target/tmp/LogisticRegressionModel"); LinearRegressionModel sameModel = LinearRegressionModel.load(sc.sc(), "target/tmp/LogisticRegressionModel"); 1.10 预测模型标记语言模型导出
spark.mllib model | PMML model |
|---|---|
| KMeansModel | ClusteringModel |
| LinearRegressionModel | RegressionModel (functionName="regression") |
| RidgeRegressionModel | RegressionModel (functionName="regression") |
| LassoModel | RegressionModel (functionName="regression") |
| SVMModel | RegressionModel (functionName="classification" normalizationMethod="none") |
| Binary LogisticRegressionModel | RegressionModel (functionName="classification" normalizationMethod="logit") |
// Load and parse the data val data = sc.textFile("data/mllib/kmeans_data.txt") val parsedData = data.map(s => Vectors.dense(s.split(' ').map(_.toDouble))).cache() // Cluster the data into two classes using KMeans val numClusters = 2 val numIterations = 20 val clusters = KMeans.train(parsedData, numClusters, numIterations) // Export to PMML println("PMML Model:/n" + clusters.toPMML) As well as exporting the PMML model to a String (model.toPMML as in the example above), you can export the PMML model to other formats: // Export the model to a String in PMML format clusters.toPMML // Export the model to a local file in PMML format clusters.toPMML("/tmp/kmeans.xml") // Export the model to a directory on a distributed file system in PMML format clusters.toPMML(sc,"/tmp/kmeans") // Export the model to the OutputStream in PMML format clusters.toPMML(System.out) 2. spark.ml:机器学习流水线高级API
2.1 概览
- DataFrame
- Transformer transform()
- Estimator fit()

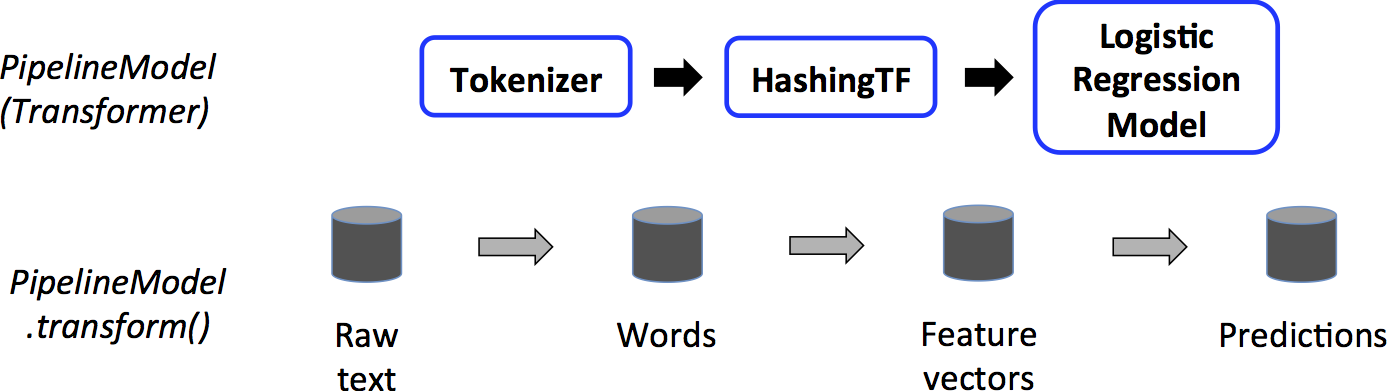
Estimator, Transformer, and Param
// Prepare training data. // We use LabeledPoint, which is a JavaBean. Spark SQL can convert RDDs of JavaBeans // into DataFrames, where it uses the bean metadata to infer the schema. DataFrame training = sqlContext.createDataFrame(Arrays.asList( new LabeledPoint(1.0, Vectors.dense(0.0, 1.1, 0.1)), new LabeledPoint(0.0, Vectors.dense(2.0, 1.0, -1.0)), new LabeledPoint(0.0, Vectors.dense(2.0, 1.3, 1.0)), new LabeledPoint(1.0, Vectors.dense(0.0, 1.2, -0.5)) ), LabeledPoint.class); // Create a LogisticRegression instance. This instance is an Estimator. LogisticRegression lr = new LogisticRegression(); // Print out the parameters, documentation, and any default values. System.out.println("LogisticRegression parameters:/n" + lr.explainParams() + "/n"); // We may set parameters using setter methods. lr.setMaxIter(10) .setRegParam(0.01); // Learn a LogisticRegression model. This uses the parameters stored in lr. LogisticRegressionModel model1 = lr.fit(training); // Since model1 is a Model (i.e., a Transformer produced by an Estimator), // we can view the parameters it used during fit(). // This prints the parameter (name: value) pairs, where names are unique IDs for this // LogisticRegression instance. System.out.println("Model 1 was fit using parameters: " + model1.parent().extractParamMap()); // We may alternatively specify parameters using a ParamMap. ParamMap paramMap = new ParamMap() .put(lr.maxIter().w(20)) // Specify 1 Param. .put(lr.maxIter(), 30) // This overwrites the original maxIter. .put(lr.regParam().w(0.1), lr.threshold().w(0.55)); // Specify multiple Params. // One can also combine ParamMaps. ParamMap paramMap2 = new ParamMap() .put(lr.probabilityCol().w("myProbability")); // Change output column name ParamMap paramMapCombined = paramMap.$plus$plus(paramMap2); // Now learn a new model using the paramMapCombined parameters. // paramMapCombined overrides all parameters set earlier via lr.set* methods. LogisticRegressionModel model2 = lr.fit(training, paramMapCombined); System.out.println("Model 2 was fit using parameters: " + model2.parent().extractParamMap()); // Prepare test documents. DataFrame test = sqlContext.createDataFrame(Arrays.asList( new LabeledPoint(1.0, Vectors.dense(-1.0, 1.5, 1.3)), new LabeledPoint(0.0, Vectors.dense(3.0, 2.0, -0.1)), new LabeledPoint(1.0, Vectors.dense(0.0, 2.2, -1.5)) ), LabeledPoint.class); // Make predictions on test documents using the Transformer.transform() method. // LogisticRegression.transform will only use the 'features' column. // Note that model2.transform() outputs a 'myProbability' column instead of the usual // 'probability' column since we renamed the lr.probabilityCol parameter previously. DataFrame results = model2.transform(test); for (Row r: results.select("features", "label", "myProbability", "prediction").collect()) { System.out.println("(" + r.get(0) + ", " + r.get(1) + ") -> prob=" + r.get(2) + ", prediction=" + r.get(3)); } Pipeline
// Labeled and unlabeled instance types. // Spark SQL can infer schema from Java Beans. public class Document implements Serializable { private long id; private String text; public Document(long id, String text) { this.id = id; this.text = text; } public long getId() { return this.id; } public void setId(long id) { this.id = id; } public String getText() { return this.text; } public void setText(String text) { this.text = text; } } public class LabeledDocument extends Document implements Serializable { private double label; public LabeledDocument(long id, String text, double label) { super(id, text); this.label = label; } public double getLabel() { return this.label; } public void setLabel(double label) { this.label = label; } } // Prepare training documents, which are labeled. DataFrame training = sqlContext.createDataFrame(Arrays.asList( new LabeledDocument(0L, "a b c d e spark", 1.0), new LabeledDocument(1L, "b d", 0.0), new LabeledDocument(2L, "spark f g h", 1.0), new LabeledDocument(3L, "hadoop mapreduce", 0.0) ), LabeledDocument.class); // Configure an ML pipeline, which consists of three stages: tokenizer, hashingTF, and lr. Tokenizer tokenizer = new Tokenizer() .setInputCol("text") .setOutputCol("words"); HashingTF hashingTF = new HashingTF() .setNumFeatures(1000) .setInputCol(tokenizer.getOutputCol()) .setOutputCol("features"); LogisticRegression lr = new LogisticRegression() .setMaxIter(10) .setRegParam(0.01); Pipeline pipeline = new Pipeline() .setStages(new PipelineStage[] {tokenizer, hashingTF, lr}); // Fit the pipeline to training documents. PipelineModel model = pipeline.fit(training); // Prepare test documents, which are unlabeled. DataFrame test = sqlContext.createDataFrame(Arrays.asList( new Document(4L, "spark i j k"), new Document(5L, "l m n"), new Document(6L, "mapreduce spark"), new Document(7L, "apache hadoop") ), Document.class); // Make predictions on test documents. DataFrame predictions = model.transform(test); for (Row r: predictions.select("id", "text", "probability", "prediction").collect()) { System.out.println("(" + r.get(0) + ", " + r.get(1) + ") --> prob=" + r.get(2) + ", prediction=" + r.get(3)); } 模型选择
// Labeled and unlabeled instance types. // Spark SQL can infer schema from Java Beans. public class Document implements Serializable { private long id; private String text; public Document(long id, String text) { this.id = id; this.text = text; } public long getId() { return this.id; } public void setId(long id) { this.id = id; } public String getText() { return this.text; } public void setText(String text) { this.text = text; } } public class LabeledDocument extends Document implements Serializable { private double label; public LabeledDocument(long id, String text, double label) { super(id, text); this.label = label; } public double getLabel() { return this.label; } public void setLabel(double label) { this.label = label; } } // Prepare training documents, which are labeled. DataFrame training = sqlContext.createDataFrame(Arrays.asList( new LabeledDocument(0L, "a b c d e spark", 1.0), new LabeledDocument(1L, "b d", 0.0), new LabeledDocument(2L, "spark f g h", 1.0), new LabeledDocument(3L, "hadoop mapreduce", 0.0), new LabeledDocument(4L, "b spark who", 1.0), new LabeledDocument(5L, "g d a y", 0.0), new LabeledDocument(6L, "spark fly", 1.0), new LabeledDocument(7L, "was mapreduce", 0.0), new LabeledDocument(8L, "e spark program", 1.0), new LabeledDocument(9L, "a e c l", 0.0), new LabeledDocument(10L, "spark compile", 1.0), new LabeledDocument(11L, "hadoop software", 0.0) ), LabeledDocument.class); // Configure an ML pipeline, which consists of three stages: tokenizer, hashingTF, and lr. Tokenizer tokenizer = new Tokenizer() .setInputCol("text") .setOutputCol("words"); HashingTF hashingTF = new HashingTF() .setNumFeatures(1000) .setInputCol(tokenizer.getOutputCol()) .setOutputCol("features"); LogisticRegression lr = new LogisticRegression() .setMaxIter(10) .setRegParam(0.01); Pipeline pipeline = new Pipeline() .setStages(new PipelineStage[] {tokenizer, hashingTF, lr}); // We use a ParamGridBuilder to construct a grid of parameters to search over. // With 3 values for hashingTF.numFeatures and 2 values for lr.regParam, // this grid will have 3 x 2 = 6 parameter settings for CrossValidator to choose from. ParamMap[] paramGrid = new ParamGridBuilder() .addGrid(hashingTF.numFeatures(), new int[]{10, 100, 1000}) .addGrid(lr.regParam(), new double[]{0.1, 0.01}) .build(); // We now treat the Pipeline as an Estimator, wrapping it in a CrossValidator instance. // This will allow us to jointly choose parameters for all Pipeline stages. // A CrossValidator requires an Estimator, a set of Estimator ParamMaps, and an Evaluator. // Note that the evaluator here is a BinaryClassificationEvaluator and its default metric // is areaUnderROC. CrossValidator cv = new CrossValidator() .setEstimator(pipeline) .setEvaluator(new BinaryClassificationEvaluator()) .setEstimatorParamMaps(paramGrid) .setNumFolds(2); // Use 3+ in practice // Run cross-validation, and choose the best set of parameters. CrossValidatorModel cvModel = cv.fit(training); // Prepare test documents, which are unlabeled. DataFrame test = sqlContext.createDataFrame(Arrays.asList( new Document(4L, "spark i j k"), new Document(5L, "l m n"), new Document(6L, "mapreduce spark"), new Document(7L, "apache hadoop") ), Document.class); // Make predictions on test documents. cvModel uses the best model found (lrModel). DataFrame predictions = cvModel.transform(test); for (Row r: predictions.select("id", "text", "probability", "prediction").collect()) { System.out.println("(" + r.get(0) + ", " + r.get(1) + ") --> prob=" + r.get(2) + ", prediction=" + r.get(3)); } DataFrame data = jsql.read().format("libsvm").load("data/mllib/sample_libsvm_data.txt"); // Prepare training and test data. DataFrame[] splits = data.randomSplit(new double[] {0.9, 0.1}, 12345); DataFrame training = splits[0]; DataFrame test = splits[1]; LinearRegression lr = new LinearRegression(); // We use a ParamGridBuilder to construct a grid of parameters to search over. // TrainValidationSplit will try all combinations of values and determine best model using // the evaluator. ParamMap[] paramGrid = new ParamGridBuilder() .addGrid(lr.regParam(), new double[] {0.1, 0.01}) .addGrid(lr.fitIntercept()) .addGrid(lr.elasticNetParam(), new double[] {0.0, 0.5, 1.0}) .build(); // In this case the estimator is simply the linear regression. // A TrainValidationSplit requires an Estimator, a set of Estimator ParamMaps, and an Evaluator. TrainValidationSplit trainValidationSplit = new TrainValidationSplit() .setEstimator(lr) .setEvaluator(new RegressionEvaluator()) .setEstimatorParamMaps(paramGrid) .setTrainRatio(0.8); // 80% for training and the remaining 20% for validation // Run train validation split, and choose the best set of parameters. TrainValidationSplitModel model = trainValidationSplit.fit(training); // Make predictions on test data. model is the model with combination of parameters // that performed best. model.transform(test) .select("features", "label", "prediction") .show(); 2.2 特征提取、转换和选择
http://spark.apache.org/docs/latest/ml-features.html
2.3 分类和回归
http://spark.apache.org/docs/latest/ml-classification-regression.html
2.4 聚类
http://spark.apache.org/docs/latest/ml-clustering.html
- 本文标签: https final map 统计 XEN Apache Hadoop mina XML swap build Word CSS root Document git REST sql core java CTO db ebay tar example IDE mmm apache parse struct list apr src rmi FIT node 实例 schema 数据 http zip HTML ip value cat lib API tab Select js explain UI ECS update App shell key find dist ACE NSA Hadoop DOM
- 版权声明: 本文为互联网转载文章,出处已在文章中说明(部分除外)。如果侵权,请联系本站长删除,谢谢。
- 本文海报: 生成海报一 生成海报二










![[HBLOG]公众号](https://www.liuhaihua.cn/img/qrcode_gzh.jpg)

